2013 continued on the new and higher level we had achieved in 2012, and continued to expand from there. We sent two people to Merrimack, we got a new Ph.D. student, we had even more people at ICSB, and we published our probably most important paper to date.
* In the spring of 2013, we did a new version of the annual course TSRT17: Systems biology modelling project. This involved around 35 students from the biomedical engineering programme (TB) and the industrial engineering biotechnology profile. As usual the projects dealt with real-life problems from different research groups, who gave the students real unanswered biological questions, which only can be answered via the usage of mathematical modelling. The students were divided into 6 projects, who worked with i) the understanding of heart desensitization, ii) insulin signalling and lipolysis in adipocytes, iii) facilitation in neurons, iv) myelin-measurements from MRI brain scans, v) estimation of liver fibrosis based on MRI time-series, vi) cell-to-cell differences in yeast cells. Group ii) did their work as a B.Sc. project. As previous years, we had a logotype-competition, and below you see a picture of the winners of their logotype.

* As usual, we recruited some of the most talented students from the course, to work in our group in various types of student projects, usually with associated scholarships. Some of these student include Philip Blomström, who worked with facilitation in neurons, and Mattias Köpsén, William Lövfors, and Fredrik Söderquist, who worked together with us, Mika Gustafsson and Mikael Benson to develop a method to estimate mechanistic ODE-models from omics data.

* We also had some older students around, and some of these were also on their way out towards their future careers. As illustrated in a previous blog entry, Linnea Bergenholm and David Janzén visited Merrimack Pharmaceuticals in Boston for a 6 month internship. This then led them to positions as Industrial Ph.D. students at AstraZeneca. This recruitment process illustrates the competitiveness of students who have been trained in our group: 2 of our students applied, both got the position. Apart from this, David Jullesson worked with the finishing of his paper, which then led him towards a Ph.D. position on Synthetic Biology with Jens Nielsen in Gothenburg. David also started up Linköping’s participation in iGEM, an international undergraduate synthetic biology competition. In this project modelling was used to some extent. Finally, Markus Karlsson worked during the entire spring semester with the application of the nonlinear mixed-effects modelling software Monolix to our liver model: this showed that our model-based approach outperforms all other image-analysis approaches, when it comes to generating biomarkers that can predict liver fibrosis.

* 2013 was also the year when a new Ph.D. student started: Mikael Forsgren (picture above). He had already been around for several years, working on modelling and experiments on the liver, and had just started a position at Wolfram MathCore, as a consultant in modelling. This Ph.D. position meant that we got him back as a 50% industrial Ph.D., and that we further strengthened our collaboration with MathCore.
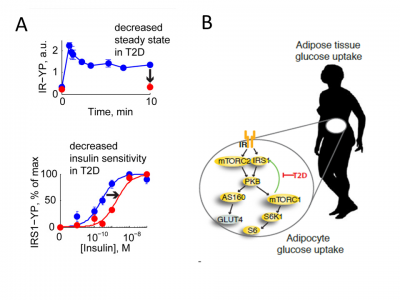 * 2013 also saw the publication of our perhaps most important publication to date: our multi-level explanation to the origin of insulin resistance. In this paper, we had complemented our existing data for insulin signalling with new experiments, to get a first collection of systems-wide time-series and dose-responses for the main players in the insulin signalling network, both normally and in type 2 diabetes (examples in sub-figure A above). These unique data allowed us to, for the first time, test existing hypotheses regarding the origin of insulin resistance, i.e. the malfunction in the insulin signalling network seen in type 2 diabetes patients. We could then show that a specific feedback, from the protein mTORC1 to the protein IRS1, is a potential such explanation. If the network is perturbed in this feedback (red arrow, sub-figure B), the entire network switches from normal to diabetic signalling. This model has since its publication gained a lot of interest from pharmaceutical companies.
* 2013 also saw the publication of our perhaps most important publication to date: our multi-level explanation to the origin of insulin resistance. In this paper, we had complemented our existing data for insulin signalling with new experiments, to get a first collection of systems-wide time-series and dose-responses for the main players in the insulin signalling network, both normally and in type 2 diabetes (examples in sub-figure A above). These unique data allowed us to, for the first time, test existing hypotheses regarding the origin of insulin resistance, i.e. the malfunction in the insulin signalling network seen in type 2 diabetes patients. We could then show that a specific feedback, from the protein mTORC1 to the protein IRS1, is a potential such explanation. If the network is perturbed in this feedback (red arrow, sub-figure B), the entire network switches from normal to diabetic signalling. This model has since its publication gained a lot of interest from pharmaceutical companies.

* Several of these showcases were presented at large international conferences, most notably the International Conference for Systems Biology (ICSB). ICSB was this year held in Copenhagen, and we went down with the largest group yet: around 15 people from at least 4 different research groups were present from Linköping. We were also well received, and Linköping had three oral presentations: Robert Palmér from the spin-off company MathCore presented our joint story on understanding the T2D drug Anakinra, Gunnar Cedersund and Markus Karlsson presented the above mentioned story on liver diagnosis, and Mikael Benson – not from our group, but also from Linköping – gave a keynote lecture on clinical end-usage of systems biology.
All in all, this year demonstrated that the previous successes of 2012 were not a one-time luck, but that we could sustain this level of activity and international visibility, and thereby bring high hopes for the coming year of 2014.

