Sundqvist N, Grankvist N, Watrous J, Mohit J, Nilsson R, Cedersund G. Validation-based model selection for 13C metabolic flux analysis with uncertain measurement errors. PLoS Comput Biol. 2022 Apr 11;18(4):e1009999. doi: 10.1371/journal.pcbi.1009999
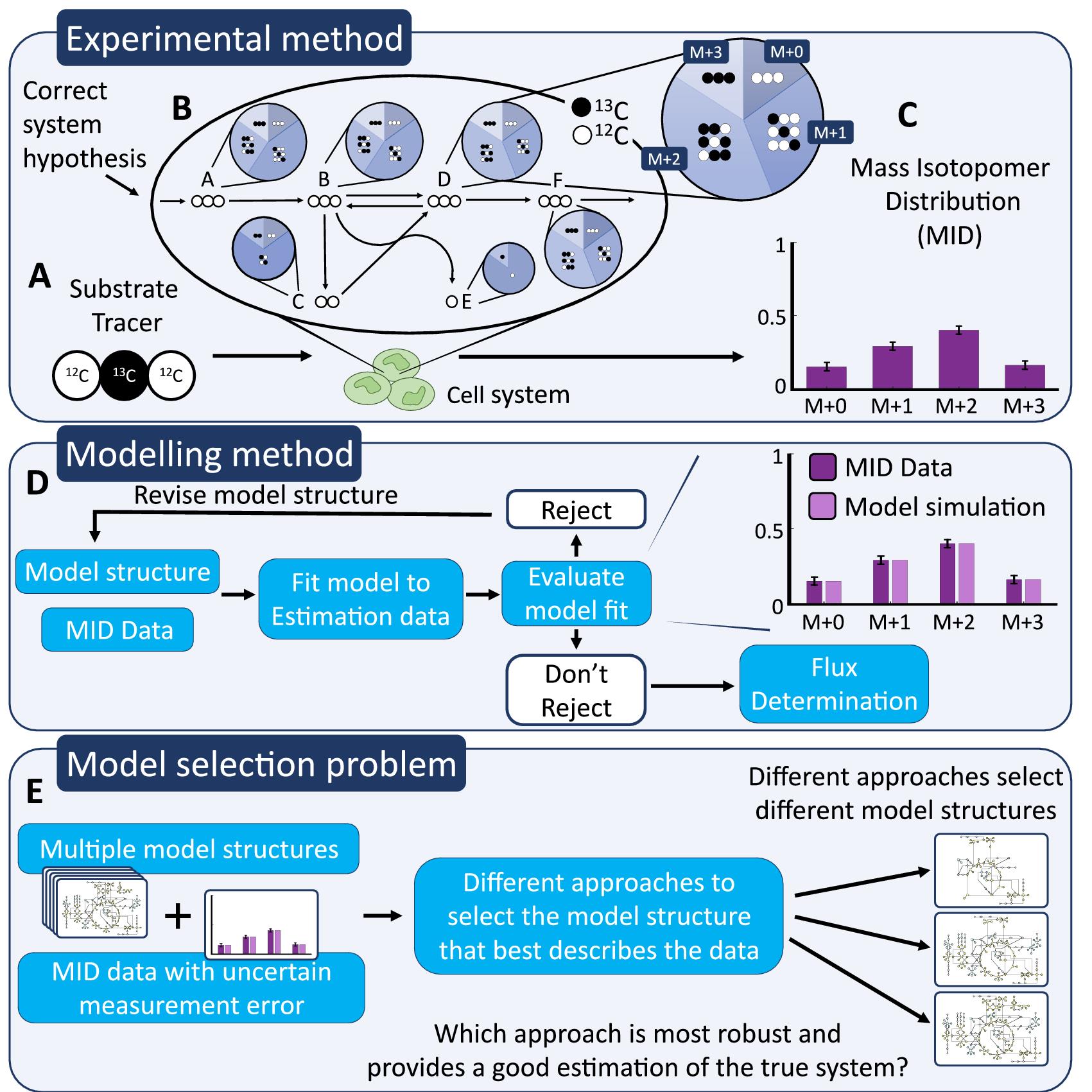
Author summary: Measuring metabolic reaction fluxes in living cells is difficult, yet important. The gold standard is to label extracellular metabolites with 13C, to use mass spectrometry to find out where the 13C-atoms ends up, and finally use mathematical modelling to calculate how quickly each reaction must have flowed, for the 13C-atoms to end up like that. This measurement thus relies on usage of the right mathematical model, which must be selected among various candidate models. In this manuscript, we present a new way to do this model selection step, utilizing validation data. Using an adopted approach to calculate the uncertainty of model predictions, we identify new validation experiments, which are neither too similar, nor too dissimilar, compared to the previous training data. The model candidate that is best at predicting this new validation data is the one chosen. Tests on simulated data where the true model is known, shows that the validation-based method is robust when the magnitude of the error in the measurement uncertainty is unknown, something that conventional methods are not. This improvement is important since true uncertainties can be difficult to estimate for these data. Finally, we demonstrate how the new method can be used on real data, to identify fluxes and important reactions.

