This year, we got a double jackpot from the Swedish Research Council – who gave us glowing reviews for the 3R project, scaling from microphysiological in vitro systems to humans using scalable digital twins.
In Sweden, the Swedish Research Council (Swe: Vetenskapsrådet, VR), is the most central research grant, and it is often considered a key quality stamp of a top researcher to have at least one grant from VR. Therefore, competition is usually fierce (acceptance rate usually is 5-15%), and it is not at all guaranteed that you get money, even if you have a competitive application. Therefore, I am proud to say that this year, I got not only one grant, but two – and that the evaluation from the reviewers was unusually high and glowing.
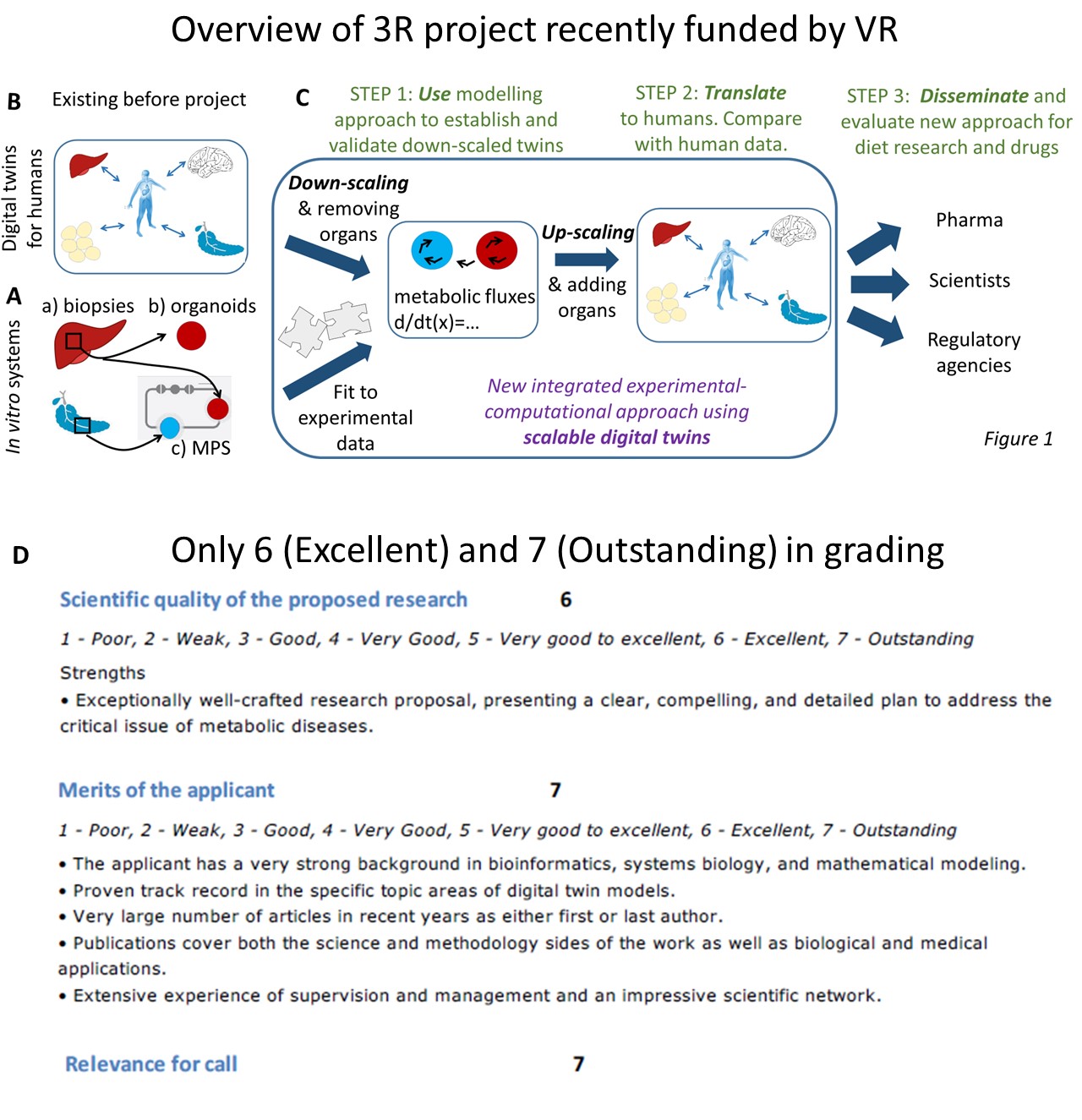
The project I have gotten the review responses for so far is a special call on 3R, i.e. Replacement, Reduction and Refinement of animal experiments. This is a topic, I have been very active in ever since 2015, when I was awarded the first edition of the prize “Nytänkaren” (the thinker of new ideas), by the Swedish Fund for Research without Animal Experiments. The project extends on our experimental work, both within the group, doing cell biology cultures using 13C-labelled metabolites on liver and adipose tissue taken from surgery), and that in collaboration with AstraZeneca, centered around organs-on-a-chip, i.e. small microphysiological systems (MPS), with organoids and spheroids consisting of human cells (Fig A, recent paper). In the project, we will i) analyse these in vitro data using mechanistic modelling to get more information out of the data (e.g. metabolic fluxes), ii) plan new experiments, by first doing the experiments in the computer, and iii) translate the results to humans, by e.g. scaling the volumes of the spheroids to human sizes, and by adding the missing organs, which allows us to re-assemble the digital twin in the computer (Fig B-C, Step 1 and 2). The project will evaluate and quantify the benefits of this for e.g. drug development, and we will disseminate the results to pharma, scientists, and regulatory agencies (Fig C, Step 3).
In the evaluation, we only got 6s and 7s, which means that we were among the highest rated of all applicants, even among the few who got money (6 out of 56). The ranking is from 1-7, where a “normal, decent” scientist usually get a 3 (meaning “good”), and where you need at least a mixture of 5s and 6s to have any chance of getting money. If you get all 6s, you are usually getting the money for sure, and 7 is only very rarely given out (I was a reviewer for ~60 applicants two years ago, and then I think only one or possibly two got a 7 on any criterion). Therefore, I am very grateful that this year, I got only 6s and higher, and that two(!) categories got a 7: “merits of applicant” and “relevance for 3R” (Fig D). If the rating levels were the same as when I was a reviewer, I would – I think – have been number one of all applicants that year, and in any case, I must have been among the very top of all the 56 applicants also this year.
The life of a scientist is filled with many many rejected applications, so when you get a jackpot once in a while, it is important to stop a bit – and celebrate! Because tomorrow, it is time to get started working on the new exciting research projects! 🙂