2012 was the year when the core facility started to take shape as a separate unit. This had been a long-time dream of mine, and when I (Gunnar Cedersund) in 2011 was awarded a VR FoAss, I could slowly start the process of building up such a joint modelling environment, working as a common knowledge and computer resource and as a physical meeting place for all groups and people wanting to do mathematical modelling of biological systems.
 The part of the group that went down to SBMC in June 2012. Above, from left: Patrick Weber, Rikard Johansson, Karin Lundengård, Mikael Forsgren, Gunnar Cedersund, Linnea Bergenholm, David Janzén, Magdalena San Roman, Elin Nyman. Front, from left: Zaheer Ali, David Jullesson.
The part of the group that went down to SBMC in June 2012. Above, from left: Patrick Weber, Rikard Johansson, Karin Lundengård, Mikael Forsgren, Gunnar Cedersund, Linnea Bergenholm, David Janzén, Magdalena San Roman, Elin Nyman. Front, from left: Zaheer Ali, David Jullesson.
Here are some of the more specific highlights:
* During the spring and summer, we had two external visitors: Patrick Weber from Stuttgart, who came to experience work in applied research and to contribute with his knowledge about Bayesian methods for parameter estimation, and Magdalena San Roman from Montevideo, Uruguay, who came to learn our type of modelling, but also to share her knowledge about metabolic control systems. They are both depicted on pictures below, and they both entered into different ongoing projects here, including in a joint project on new improved methods for sensitivity analysis with uncertainty.
 Patrick Weber, our visitor from Stuttgart, Germany
Patrick Weber, our visitor from Stuttgart, Germany
* During the spring, Eva-Maria Hansson, who had previously worked and done her M.Sc. thesis with us, went to visit Merrimack Pharmaceuticals in Boston, US. She was therefore the first student, who initiated a tradition and collaboration between SBCF and this company: we train really talented students in the wonderful art of systems biology, and when they are finished, we each year send some 1-3 of these students to work in internships for them. These internships are payed employements where you do modelling in pairs with an experimentalist at Merrimack, together with which you test out a new idea for a drug or a biological mechanism. If the project turns out to be promising, they will take it further, and – with some luck – the results will one day form the basis for a new drug or treatment.

* During the year, we also had some internal M.Sc. students who finished their projects for us. For instance, Linnea Bergenholm, who had already been working for a year with us prior to her thesis (mentioned here), and Zaheer Ali, who initiated the work on facilitation, that we presented in Liepzig (mentioned below). Another Ph.D. student was David Janzén, who worked on usage of mixed-effect modelling for estimation of noise and for analysis of cell-to-cell variation.
* In June, we attended the bi-annual conference Systems Biology of Mammalian Cells (SBMC), in Leipzig. SBMC is probably the second biggest conference on systems biology, and the focus fits our research very well. We went down 11 people in two big rented cars/vans, and spent 4 days in what I will probably for a long time consider as the defining trip – as the trip that defines the ultimate group trip. That was not only because we were a number of people who had started to grow together over the years (see the picture above), but also because we made such a big overall impression on the event: we had 3 oral presentations, and I was chair for one of the sessions. This probably made us the most well-represented group at the whole event! The three talks are available as videos, and were given by Elin Nyman (who spoke about her recent paper on insulin signalling in rodents), Karin Lundengård (who spoke about modelling to unravel the mechanisms behind facilitation in neurons), and Mikael Forsgren (who spoke about our clinically useful modelling on liver diagnosis).
 Our guest Magdalena San Roman, who took a beer at SBMC together with Jan Hasenauer, who was a previous guest of ours, from a few years earlier.
Our guest Magdalena San Roman, who took a beer at SBMC together with Jan Hasenauer, who was a previous guest of ours, from a few years earlier.
* In August 2012, I also – at last – published a paper on the set of methods for core prediction analysis that I had presented on posters and presentations for several years (it was also an invitated paper, following my talk on ICSB the year before). This was the presumably most important paper of that year, and it forms the basis for many future systems biology papers, both on methods and on applications in various fields. However, some other papers worth mentioning are this paper on describing large-scale networks in a manner consistent with modelling (developed together with our guest professor, Hiroaki Kitano), this review on modelling of insulin signalling, and the insulin signalling paper Elin Nyman presented at SBMC (video link above).
* Following these developments done during the first half of the year, on Sept 24 2012, we were finally ready to declary ourselves as an independent group. This date is henceforth referred to as the SBCF day, the day our systems biology core facility was officially launched.
 Picture of Karin Lundengård, our new Ph.D. student.
Picture of Karin Lundengård, our new Ph.D. student.
* 2012 was also the year when Karin Lundengård started her Ph.D. with us. In practice she had been around since her M.Sc. thesis about a year earlier, but now we had gone through all the steps of finding money, supervisors, and the formal process of announcing positions etc. Karin is having me as co-supervisor: her main supervisor is Maria Engström, and her other co-supervisor is Fredrik Elinder. Fredrik has experience of both modelling and experiments on neuronal cell-biology and electrophysiology, and Maria is an expert on fMRI, used to investigate brain activity. Karin will in her thesis combine these levels, and provide a more mechanistic understanding for the data that can be obtained from fMRI, and thereby get more information from the data, eventually leading to better usage of the technique, and to better understanding and treatment of diseases like narcolepsi.
 In Dec of 2012, Cecilia Brännmark graduated from her Ph.D. studies. Here we see her during the ritual known as “spikning”, where she nails her thesis to the “thesis tree”, seen to the right.
In Dec of 2012, Cecilia Brännmark graduated from her Ph.D. studies. Here we see her during the ritual known as “spikning”, where she nails her thesis to the “thesis tree”, seen to the right.
* At the other end of the spectrum, 2012, was also the year when our first Ph.D. student graduated, i.e. finished her Ph.D. studies. The student, Cecilia Brännmark, was the first one I (Gunnar) supervised, and she started back in the days when I did all the modelling myself. Hence, Cecilia was an experimental Ph.D. student, and she has done the majority of the experiments in the early defining papers, that came out of my collaboration with prof Peter Strålfors, who was her main supervisor.
* In December we arranged a 10-day Ph.D. course on Multi-level modelling of gluocse homeostasis. During these 10 days we had invited the most relevant and world-leading experts on systems biology research on all the main organs: the liver, fat tissue, skeletal muscle, the brain, the pancreas, and the whole-body level. For all these organs we had invited experts on both the experimental research and on the modelling.

* As a separate event in the middle of this course, we also arranged a workshop on Systems Medicine. This was a co-arrangement with Mikael Benson, who arranged the first day devoted to omics and large-scale network approaches, and isbgroup/me, who arranged the second day devoted to modelling. In other words, we tried to bring together all the different sides that are relevant to systems medicine, and regarding the modelling this importantly included the neighbouring biomodelling fields, such as pharmacokinetic/pharmacodynamic modelling, image-based modelling of the liver, medical decision making with probabilistic graphical models, and even an overview of the neighbouring research in technical diagnosis. At the event there was also a strong presence from several leading drug development companies. The event was atteneded by some 60-80 people, and the combined 10-day event was the – by far – biggest arrangement we had arranged at that point.






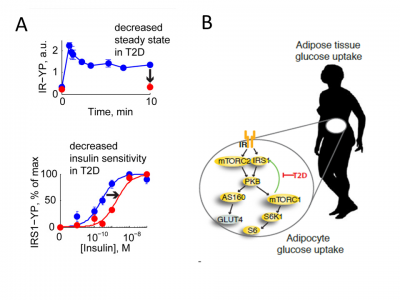 * 2013 also saw the publication of our perhaps most important
* 2013 also saw the publication of our perhaps most important 
 The part of the group that went down to SBMC in June 2012. Above, from left: Patrick Weber, Rikard Johansson, Karin Lundengård, Mikael Forsgren, Gunnar Cedersund, Linnea Bergenholm, David Janzén, Magdalena San Roman, Elin Nyman. Front, from left: Zaheer Ali, David Jullesson.
The part of the group that went down to SBMC in June 2012. Above, from left: Patrick Weber, Rikard Johansson, Karin Lundengård, Mikael Forsgren, Gunnar Cedersund, Linnea Bergenholm, David Janzén, Magdalena San Roman, Elin Nyman. Front, from left: Zaheer Ali, David Jullesson.  Patrick Weber, our visitor from Stuttgart, Germany
Patrick Weber, our visitor from Stuttgart, Germany
 Our guest Magdalena San Roman, who took a beer at SBMC together with Jan Hasenauer, who was a previous guest of ours, from a few years earlier.
Our guest Magdalena San Roman, who took a beer at SBMC together with Jan Hasenauer, who was a previous guest of ours, from a few years earlier.  Picture of Karin Lundengård, our new Ph.D. student.
Picture of Karin Lundengård, our new Ph.D. student.  In Dec of 2012, Cecilia Brännmark graduated from her Ph.D. studies. Here we see her during the ritual known as “spikning”, where she nails her thesis to the “thesis tree”, seen to the right.
In Dec of 2012, Cecilia Brännmark graduated from her Ph.D. studies. Here we see her during the ritual known as “spikning”, where she nails her thesis to the “thesis tree”, seen to the right. 
