So, now
vacation time is over here in Sweden – and work is back with its full
hustle-bustle frenzy. I (Gunnar) started working on Wednesday last week, and
since then I have already submitted one application (to SSF’s industrial Ph.D. programme together with AstraZeneca,
on the creation of a new systems pharmacology platform, based on modelling which is used to
bridge between pre-clinical organs-on-a-chip data and human/clinical
studies), had two supervision days, changed office (my main office is now at
the Department of Biomedical Engineering, and not Clinical and Experimental
Medicine), did the final preparations for the half ironman race this weekend, and went to
Gothenburg to give a lecture in the group of Patrik Rorsman and Charlotta
Olofsson. It is about this last topic, my presentation, that I want to say a
few extra words also here.
 Figure 1: Front page of my presentation earlier today.
Figure 1: Front page of my presentation earlier today.
The title
of my presentation was “Doing biology without modelling is like driving without
a safety belt – it might work, but it might also go really, really wrong”. The
talk was a salespitch to an essentially all experimentalist-audience, and it is
based on an image that came to my mind just a few days ago. The image is a response
to some of the most common forms of critique I usually hear against the usage
of modelling: “I could have said that without using the model”, “I don’t believe
that a model can do everything, I think there is still too much that we don’t
understand”, and “the model only seems to provide an extra degree of confidence
on a conclusion I would have said anyway”.
And, the
thing is that I actually – to a large extent – agree with all of those
statements. Modelling cannot do everything, and should not be oversold – but it can do some things, and those things should be properly appreciated. Similarly, many modelling results are conclusions one could have drawn without
the usage of modelling, and what the modelling does is therefore in many respects primarily
to put and extra degree of confidence on that conclusions, if that was the conclusion you anyway would have made. And just like for a safety
belt, when driving: often you don’t need it, you would be fine anyway, but it
is more secure to have it there, to bring an extra degree of security and
confidence to the current situation. In other words, if a model agrees with
your conclusion, you can be more sure that you are correct. However, in the
presentation, I also gave several examples of cases where the modelling
provides a conclusion that seems perfectly reasonable once you see it, but
where the prevailing conclusion before the model-analysis was done, actually
was a very different one. Apart from our main diabetes examples, I pointed to three probably less known such stories:
 Figure 2: The difference between the new way of calculating EC50 values (black) with the old one (red).
Figure 2: The difference between the new way of calculating EC50 values (black) with the old one (red).
1) Our EC50
story, published in FEBS J last year. There we showed that a simple model-analysis could detect a problem with
a previous way of calculating EC50 values: that the steady-state was not
reached in-between the changes in stimuli. In other words, the resulting curve was not an increase in steady-state values, but a long transient overlaying of overshoot responses. That
new interpretation of the data had some predictions, which we verified in independent
experiments, supporting our new interpretation of the data. Furthermore, it was
experimentally not possible to modify the protocol, to wait as long as one should to reach steady-state. Therefore,
the only way for this system to get correct EC50 values (corresponding to steady-state values) is to do what we
propose: to fit the model to a transient responses like the one already existing, and
then use the model to simulate the experiment as it ideally should have been done to start with. As you see in Figure 2 above, the new, more correct EC50 value is almost completely non-overlapping with the old one.
2) An
earlier story of muscle metabolism, where we showed that a seeming
contradiction and missing link – which had been investigated for 25 years – in fact
was not a contradiction at all, but merely a mis-interpretation of data. And
that re-interpreted version sounds very reasonable once you see it. In other
words, for 25 years, people had believed that a not yet discovered regulator of
glycolysis was active in anaerobic muscle recovery, but our modelling showed
that no search for such an unknown regulator is necessary: a correct analysis
of the data shows that the conventional regulators are sufficient to explain
the observations. This story is not yet published, but anyway available as
chapter 11 in my Ph.D. thesis.
3) Another
recent story on interpretation of data for the IL1beta analog Anakinra (Palmér et al, CPT Pharmacometrics Syst Pharm, 2014). In this
story, we had a look at data that seemed to be too good to be true, and
therefore were disbelieved by many: that Anakinra could have lasting positive
effects on the diabetes readout HbA1c as long tie as 1 year after the start of
the treatment, even though the treatment itself only lasted for 3 months. We
showed that a simple model based on pre-clinical data alone did actually
produce that clinical output as well. In other words, we showed that the
initial response was that the initial disbelief in the clinical data was unnecessary: they were perfectly aligned with the pre-clinical data.
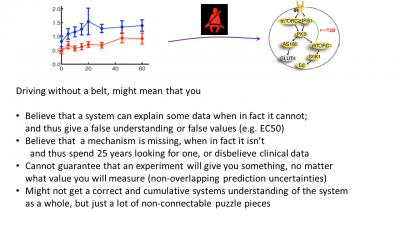 Figure 3: Last page in the presentation, summing up the main advantages of doing modelling, i.e. pointing out some of the drawbacks of not using it.
Figure 3: Last page in the presentation, summing up the main advantages of doing modelling, i.e. pointing out some of the drawbacks of not using it.
These are three examples that show that
modelling in principle does not do anything that one anyway does: analysis
of experimental data to draw conclusions and suggest new experiments. However, modelling does these things in a more reliable fashion, and it is easy to go wrong otherwise. In other words, to do biological data
analysis without modelling, is just like to driving without a safety belt: it
might work, but it might also go really really bad – throwing away 25 years
of your life.
 Figure 4: On the train on the way back, I happened to be seated right next to a very nice systems biology colleague of mine: Adil Mardinoglu. He told me that he and some colleagues of him had read my last blog post, and wanted to contact me about it. So that is actually the reason why I got inspired to write a new one as well!
Figure 4: On the train on the way back, I happened to be seated right next to a very nice systems biology colleague of mine: Adil Mardinoglu. He told me that he and some colleagues of him had read my last blog post, and wanted to contact me about it. So that is actually the reason why I got inspired to write a new one as well!
 Gunnar defending his thesis 2006.
Gunnar defending his thesis 2006. ISB group 2010.
ISB group 2010. ISB group 2012.
ISB group 2012. ISB group 2015.
ISB group 2015.
 Figure 1: Me giving a lecture, before recieving the award Nytänkaren from the Swedish Fund for Research without Animal Experiments.
Figure 1: Me giving a lecture, before recieving the award Nytänkaren from the Swedish Fund for Research without Animal Experiments.  Figure 2: The new report just issued by FDA. Note the date, it was published a few weeks ago!
Figure 2: The new report just issued by FDA. Note the date, it was published a few weeks ago! Figure 3: First page of my keynote presentation today. It is one of the first times, perhaps even the first time, that I am giving a lecture denoted as keynote, so I am looking much forward to it!
Figure 3: First page of my keynote presentation today. It is one of the first times, perhaps even the first time, that I am giving a lecture denoted as keynote, so I am looking much forward to it! Figure 1: Front page of my presentation earlier today.
Figure 1: Front page of my presentation earlier today.  Figure 2: The difference between the new way of calculating EC50 values (black) with the old one (red).
Figure 2: The difference between the new way of calculating EC50 values (black) with the old one (red).  Figure 3: Last page in the presentation, summing up the main advantages of doing modelling, i.e. pointing out some of the drawbacks of not using it.
Figure 3: Last page in the presentation, summing up the main advantages of doing modelling, i.e. pointing out some of the drawbacks of not using it.  Figure 4: On the train on the way back, I happened to be seated right next to a very nice systems biology colleague of mine: Adil Mardinoglu. He told me that he and some colleagues of him had read my last blog post, and wanted to contact me about it. So that is actually the reason why I got inspired to write a new one as well!
Figure 4: On the train on the way back, I happened to be seated right next to a very nice systems biology colleague of mine: Adil Mardinoglu. He told me that he and some colleagues of him had read my last blog post, and wanted to contact me about it. So that is actually the reason why I got inspired to write a new one as well!  A graphical depiction of the model of all added proteins. The old model we had previously developed is depicted in yellow, all proteins that are connected by high-confidence interactions (3 or more references) are depicted in dark blue, and all proteins reached using one or several low-confidence interactions are depicted in light blue. All proteins can describe time-course data.
A graphical depiction of the model of all added proteins. The old model we had previously developed is depicted in yellow, all proteins that are connected by high-confidence interactions (3 or more references) are depicted in dark blue, and all proteins reached using one or several low-confidence interactions are depicted in light blue. All proteins can describe time-course data.  Picture of William, also used on his
Picture of William, also used on his 





 TITLE AND ABSTRACT
TITLE AND ABSTRACT
